|
News
- [November, 2020] - Joined CSM as a Research Scientist.
- [October, 2020] - Our paper on meshing the output points of a point network accepted to 3DV 2020.
- [August, 2020] - Our paper on style compatibility of 3D furnitures accepted to Pacific Graphics 2020.
- [March, 2020] - Gave a 3hr talk (slides available) on 3D Deep Learning @ UCI.
- show more
- [August, 2019] - I received the travel award to attend BMVC 2019.
- [July, 2019] - Our paper on Learning Embedding of 3D models with Quadric Loss accepted for Oral
Presentation (acceptance rate - 4.6%) at BMVC 2019.
- [April, 2018] - Our paper on Processing Mouse Brain Slices accepted to Journal of Neuroscience Methods. We have also released our code and data with the project.
- [January, 2018] - Our paper on Transcribing Tiny-Occuluded Labels is accepted to WACV 2018.
- [November, 2017] - I received the prestigious ICS Innovation Endowed Fellowship for 2018.
- [May, 2017] I received Best Demo Award (video) @ UCI Computer Science Research Showcase 2017.
- [December, 2016] I received the prestigious Public Impact Fellowship for 2016-2017
- [July, 2016] Our paper on Automatic Artifact Detection is accepted for Long Oral Presentation to MICCAI MCV Workshop 2016.
|
|
Research
I enjoy solving problems at the intersection of computer vision, graphics and 3D deep learning. In the past, I have also worked on low-level vision, medical image processing and 3D visualization.
|
 |
High-Quality 3D Models (Coming Soon)
High quality mesh dataset comprising of 3D models which contain artistic embellishments and decorations.
|
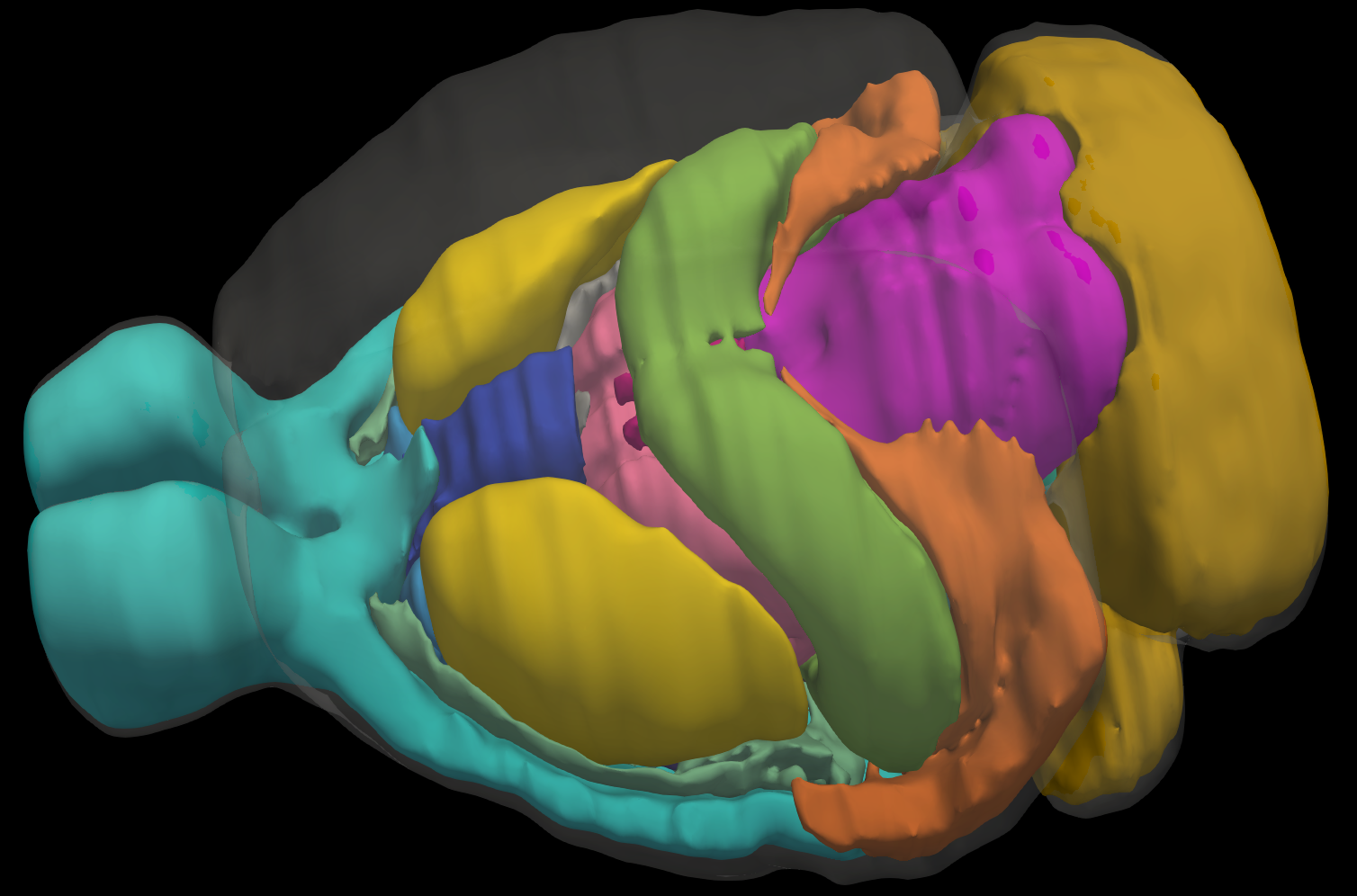 |
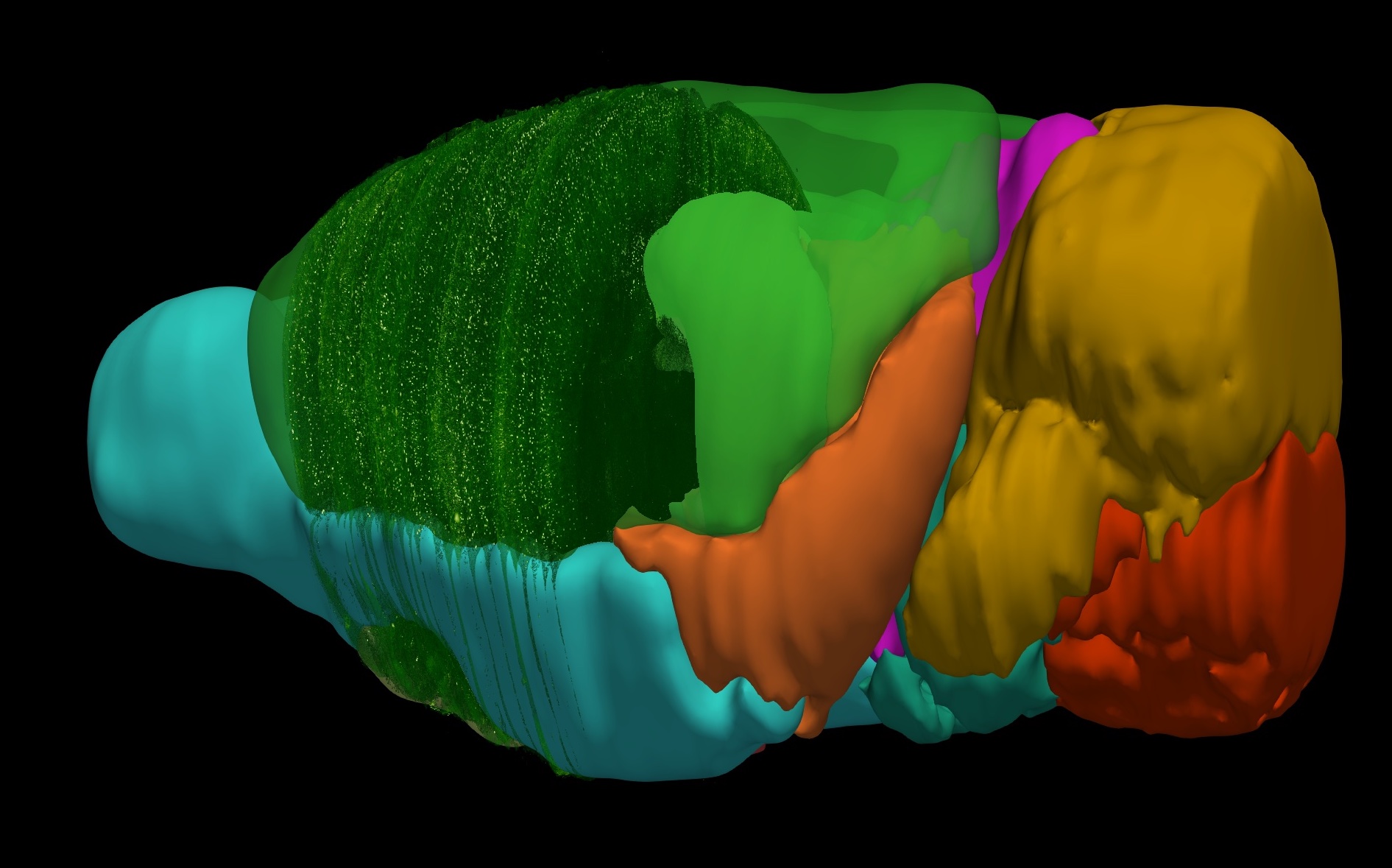
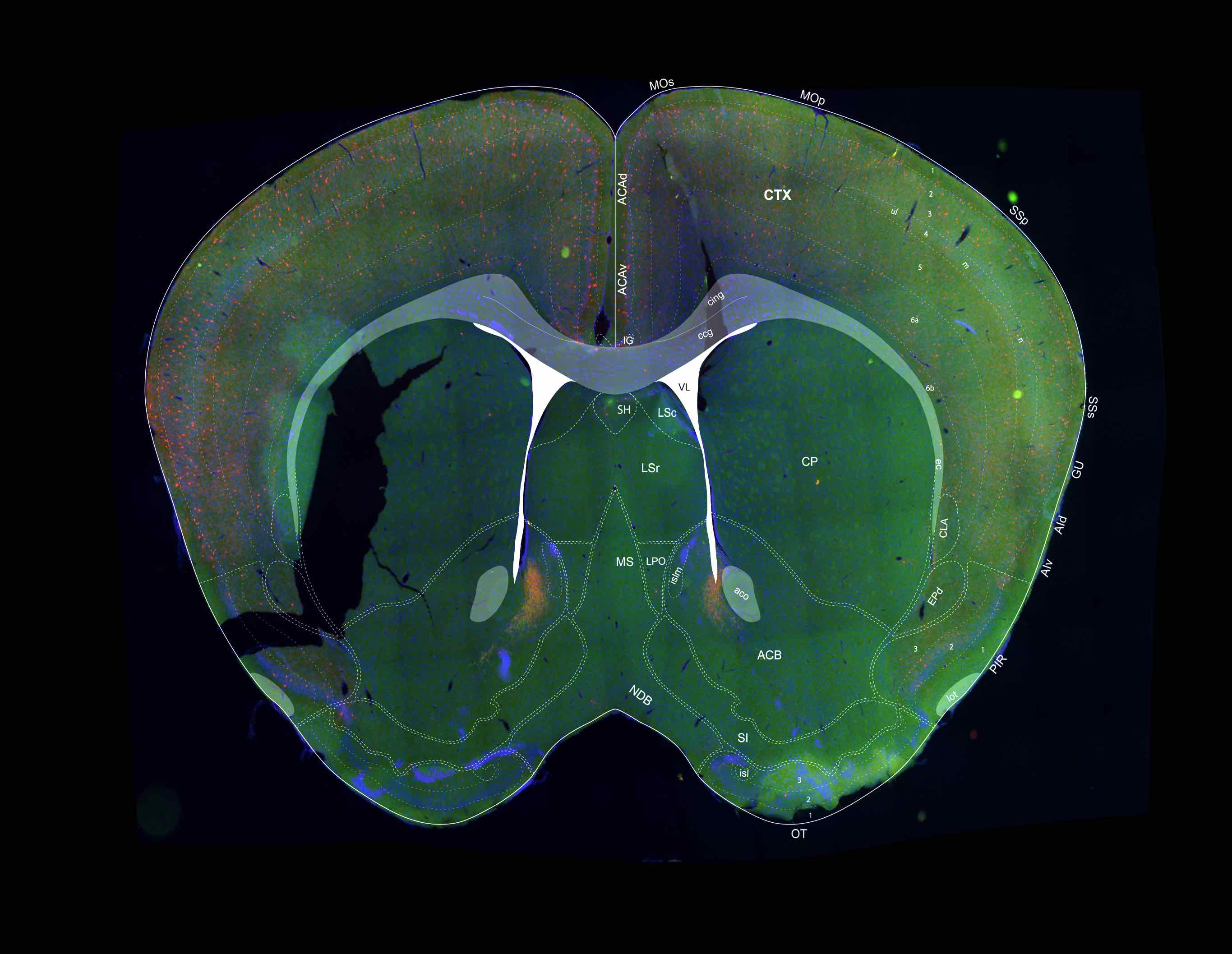
3D Annotated Mouse Brain Model
A 3D annotated mouse brain model constructed from the 2D Allen Reference Atlas. Our 3D model has correct topology and geometry and can be virtually sliced in arbritary directions and intervals serving as a template for automatic registration.
v1 - with 20 major anatomical regions
v2 - with 166 major anatomical regions (Coming Soon)
|
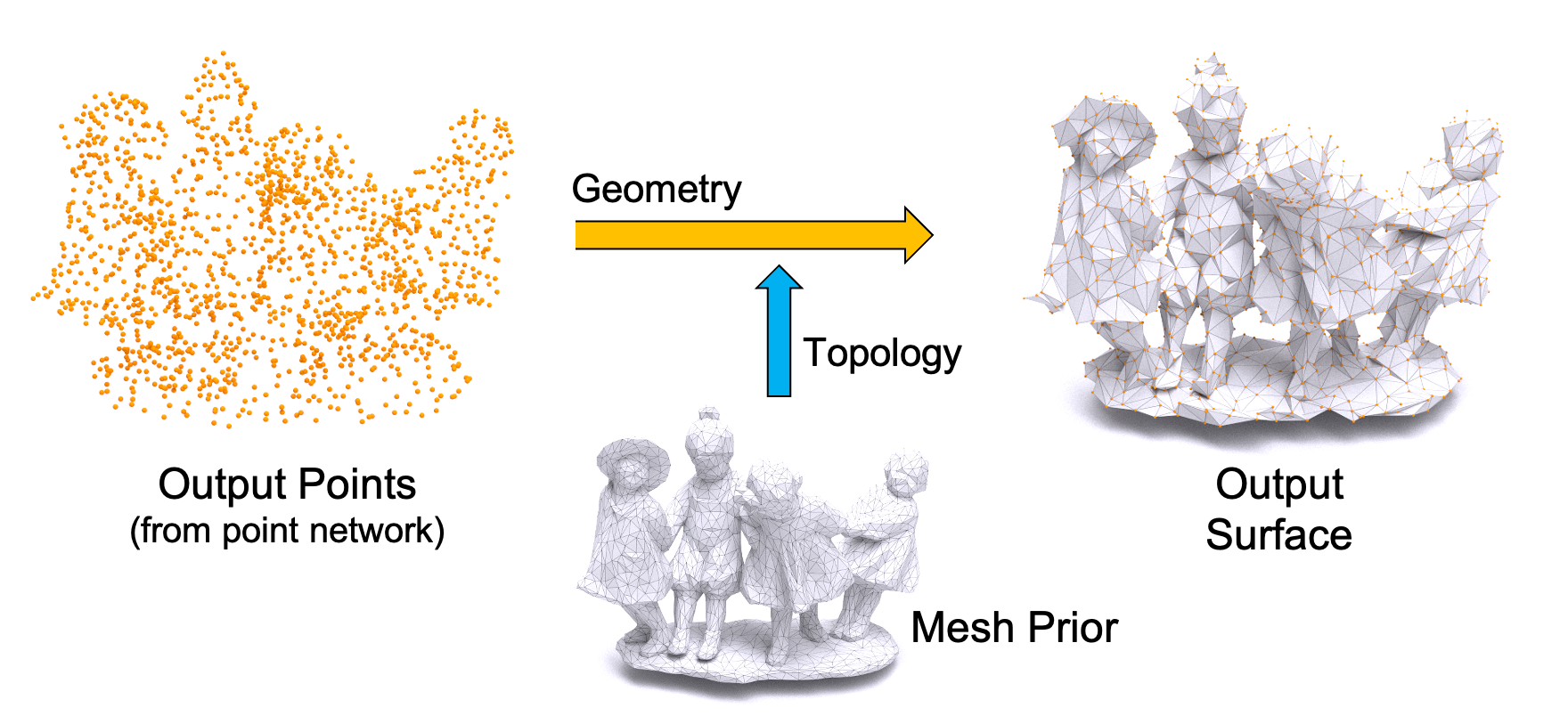 |
GAMesh: Guided and Augmented Meshing for Deep Point Networks
Nitin Agarwal, Gopi Meenakshisundaram
International Conference on 3D Vision (3DV), 2020
project page /
poster /
bibtex
We propose a meshing algorithm to generate a surface with correct topology for the output points of a point network. GAMesh can be used both in post-processing to mesh the output points or to train the point network to directly optimize the vertex positions of the final 3D mesh.
|
 |
Image-Driven Furniture Style for Interactive 3D Scene Modeling
Tomer Weiss, Ilkay Yildiz, Nitin Agarwal, Esra Ataer-Cansizoglu, Jae-Woo Choi
Pacific Graphics, 2020
paper /
bibtex /
video
We infer the style of 3D furniture models by training a variation of siamese network on scene images. Our style estimation not only embodies geometry, but also other elements reflected in scene images inlcuding color, texture, material, illumination and the use of space.
|
 |
Tutorial on 3D Deep Learning
Nitin Agarwal
Course on Advanced Computer Graphics, Winter 2020
slides
I gave a 3hr talk on 3D deep learning in the graduate class (CS-211B) taught by Prof. Gopi Meenaskshisundaram at UCI, where I provided an overview of the area along with a lot of resources useful for anyone starting research in this direction.
|
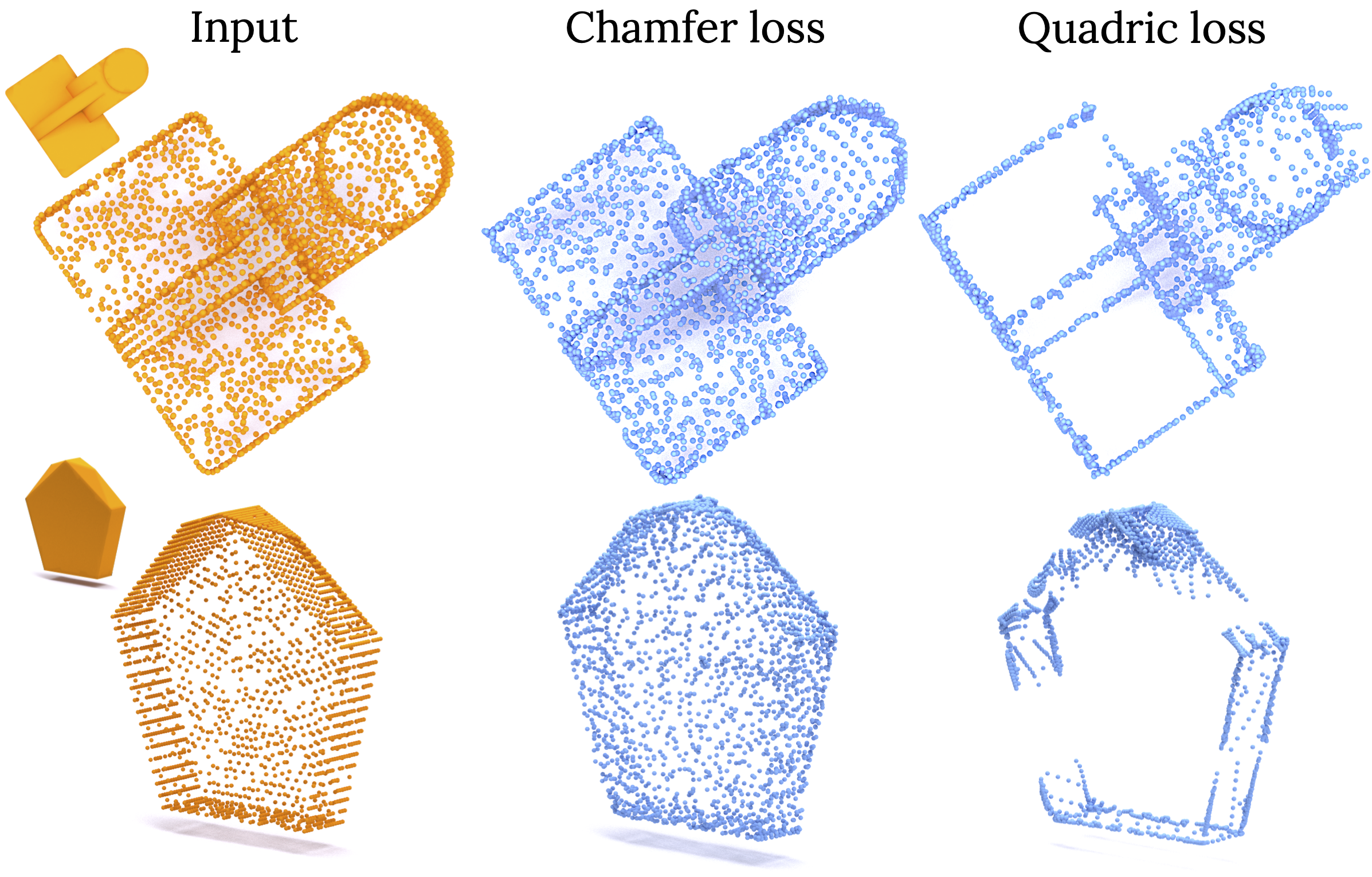 |
Learning Embedding of 3D models with Quadric Loss
Nitin Agarwal, Sung-Eui Yoon, Gopi Meenakshisundaram
British Machine Vision Conference (BMVC), 2019 (Oral Presentation)
project page /
slides /
poster /
bibtex
We propose a new point-to-surface based loss function named Quadric Loss, which penalizes displacements of points in the normal direction thereby preserving sharp features and edges in the output reconstruction. Its differentiable and can be easily incorporated into any point/mesh based network.
|
|
Multi-View Geometry & 3D Reconstruction
|
|
Low Level Vision, Computer Graphics & 3D Visualization
|
 |
Geometry Processing of Conventionally Produced Mouse Brain Slice Images
Nitin Agarwal, Xiangmin Xu, Gopi Meenakshisundaram
Journal of Neuroscience Methods, 2018
project page /
bibtex
We develop techniques and algorithms for automatic registration and 3D reconstruction of conventionally produced mouse brain slices in a standardized atlas space.
|
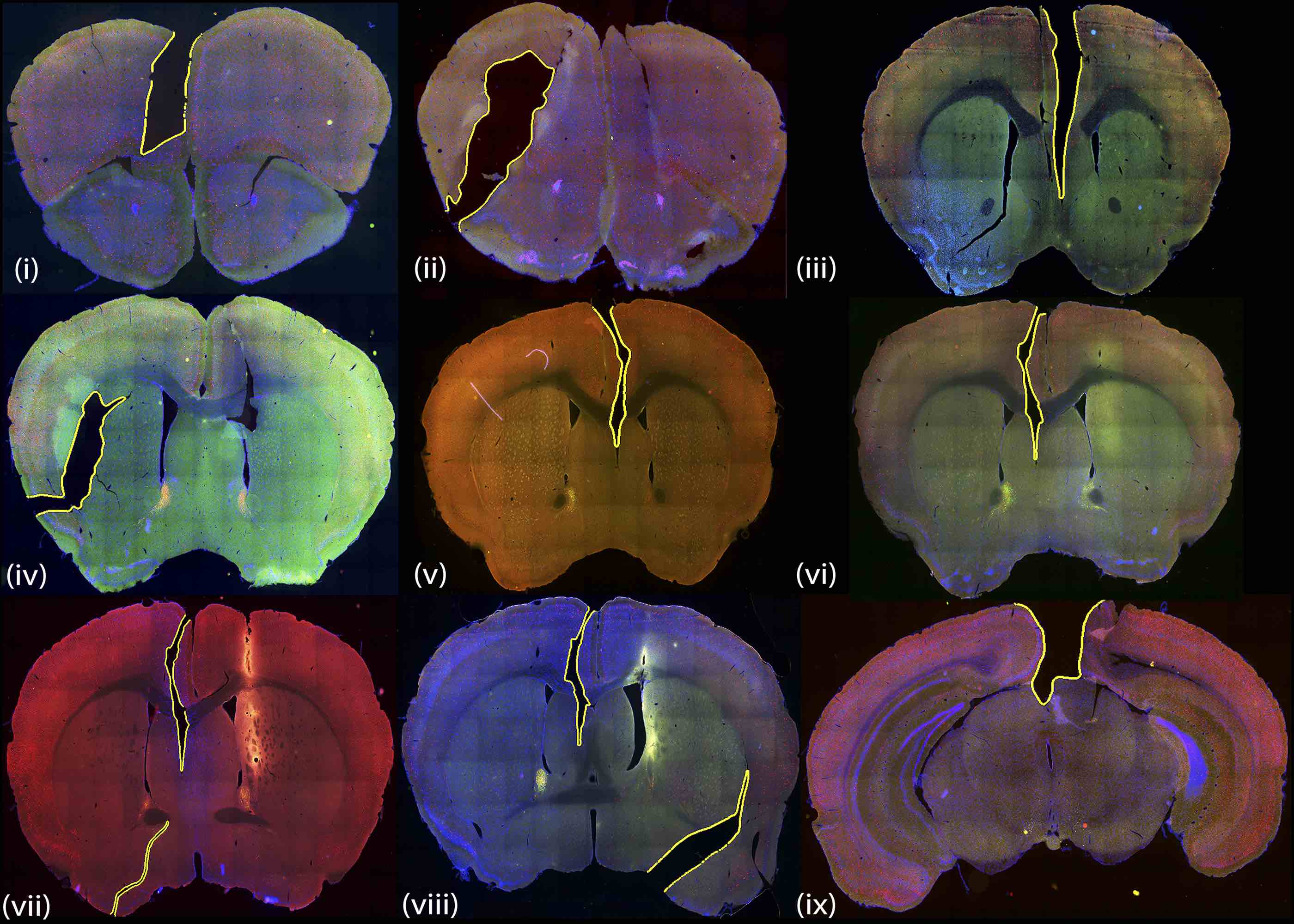 |
Automatic Detection of Histological Artifacts in Mouse Brain Slice Images
Nitin Agarwal, Xiangmin Xu, Gopi Meenakshisundaram
Workshop on Medical Computer Vision: Algorithms for Big Data (MICCAI), 2016 (Oral Presentation)
project page /
slides /
poster /
bibtex
Histological artifacts are extremely common in conventional histological procedures. In this work we present a new method to automatically detect and ignore such artifacts for achieveing accurate registration.
|
 |
Robust Registration of Mouse Brain Slice with Severe Histological
Artifacts
Nitin Agarwal, Xiangmin Xu, Gopi Meenakshisundaram
Indian Conference on Computer Vision, Graphics & Image Processing (ICVGIP), 2016
project page /
poster /
bibtex
We propose a method for non-linear registration of mouse brain histology slices (with various histological artifacts) to a standardized atlas space.
|
|
|
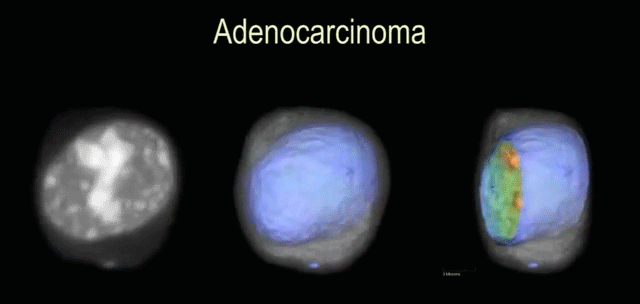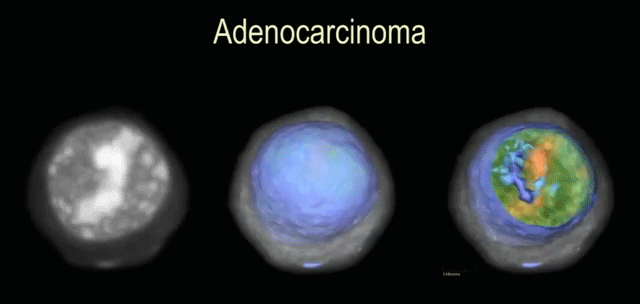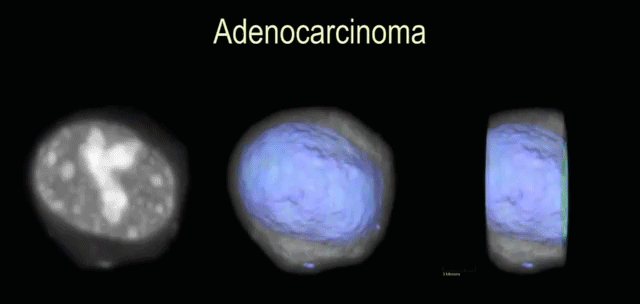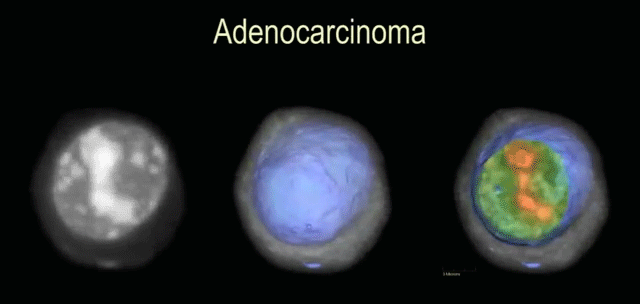
DNA ploidy measure of Feulgen-stained cancer cells using 3D image cytometry
Nitin Agarwal, Yiting Xie, Florence Patten, Anthony P. Reeves, Eric Seibel
IEEE Engineering in Medicine and Biology Society (EMBS), 2014 (Oral Presentation)
project page /
slides /
bibtex
A method to accurately measure the DNA content of single cells in 3D and show that it can be an alternative method for assessing tumorgenesis in diagnostic cells.
|
|
|
3D DNA image cytomtery by optical projection tomographic microscope for early cancer diagnosis
Nitin Agarwal, Alberto Biancardi, Florence Patten, Anthony P. Reeves, Eric Seibel
Journal of Medical Imaging (JMI), 2014
project page /
bibtex
We extend the current cytopathological techniques from 2D to 3D for early cancer detection. We do this by accurately imaging individual cells along with novel 3D nuclear segmentation algorithms.
|
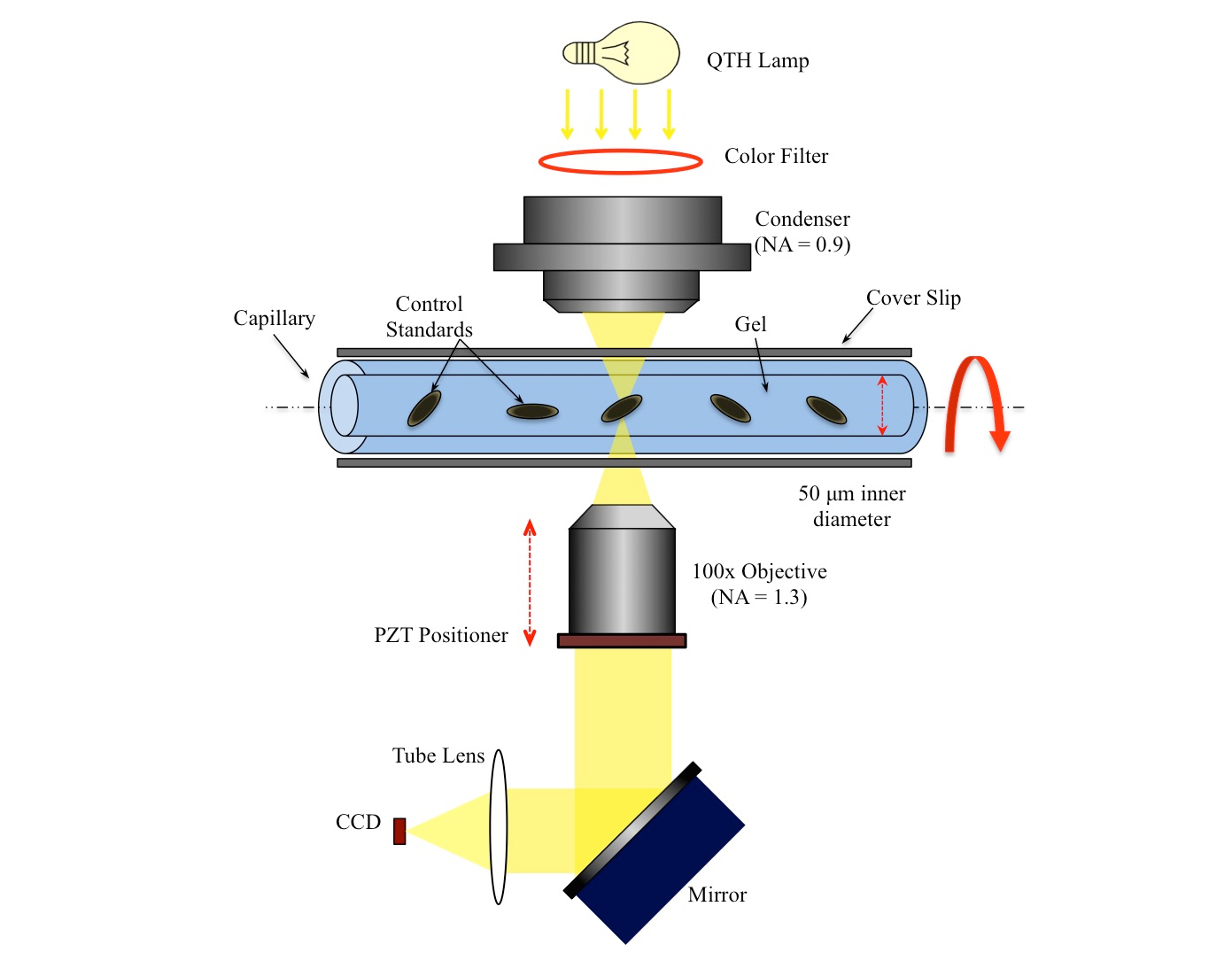 |
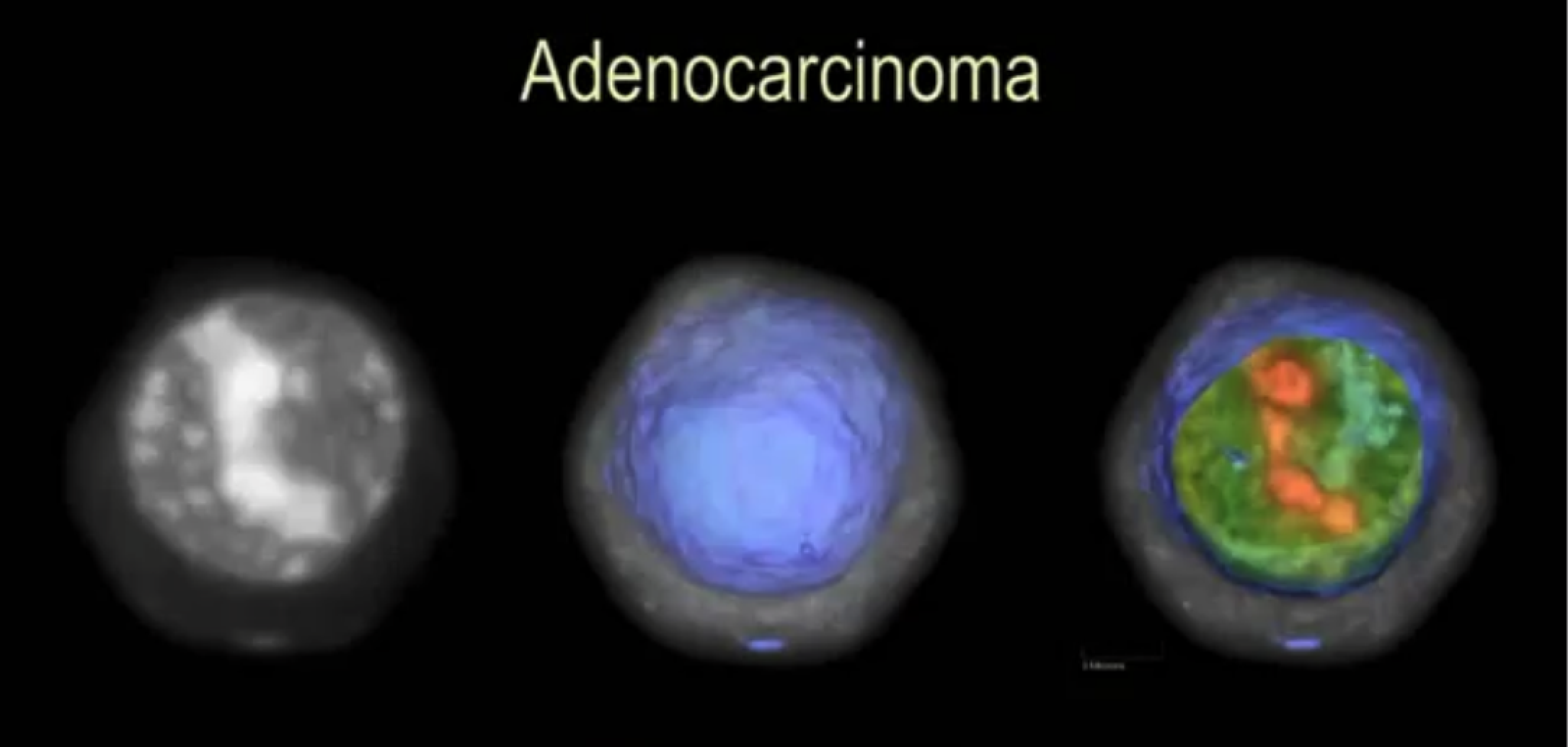
Quantification of relative chromatin content in flow cytometry standards using 3D OPTM imaging technique
Nitin Agarwal, Alberto Biancardi, Florence Patten, Anthony P. Reeves, Eric Seibel
SPIE Medical Imaging (SPIE), 2013
project page /
poster /
bibtex
A method to accurately measure the DNA content in small Flow Cytometry standards (chicken and trout nuclei).
|
 |
Conferences & Journals Reviewed
Vision & Graphics: Graphical Models (2020), Pacific Graphics (2019), TVCG, I3D (2015), Visual Computer.
Medical Image Processing: PLOS ONE, Bioinformatics, Neuroscience Methods, Methods in Ecology & Evolution.
|
|
Personal
|
|
|
In my spare time I love to do photography. Do check out my images.
|
|