|
|
|
|
|
|
|
|
|

|
|

|
|
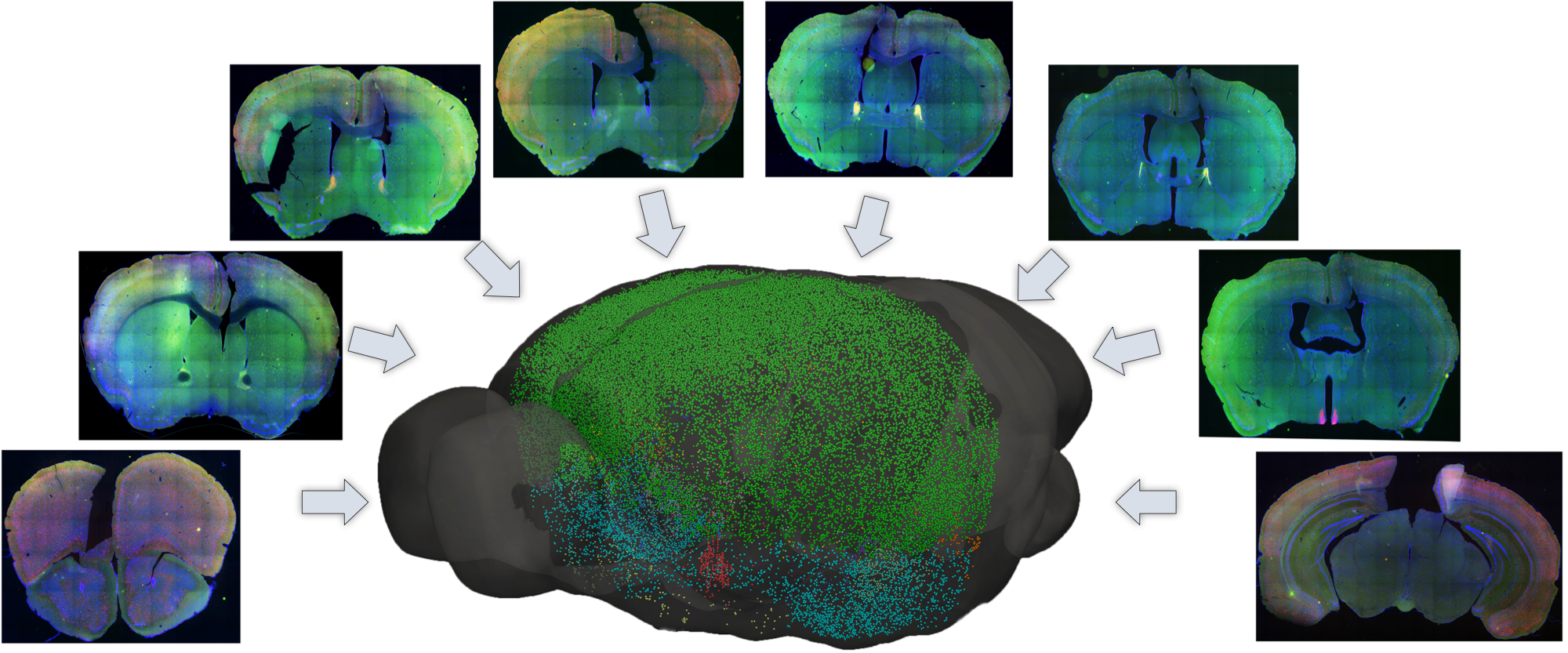
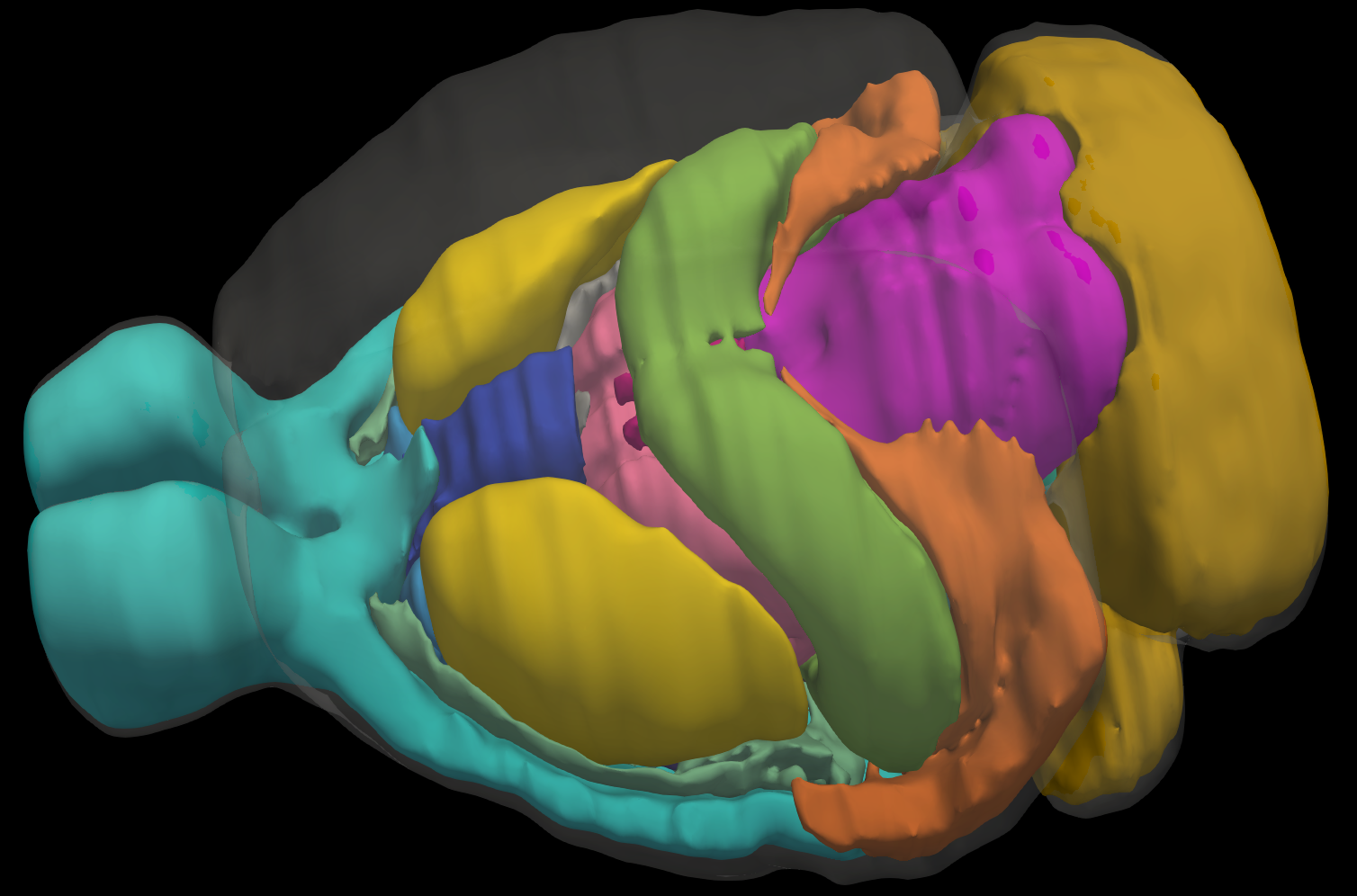
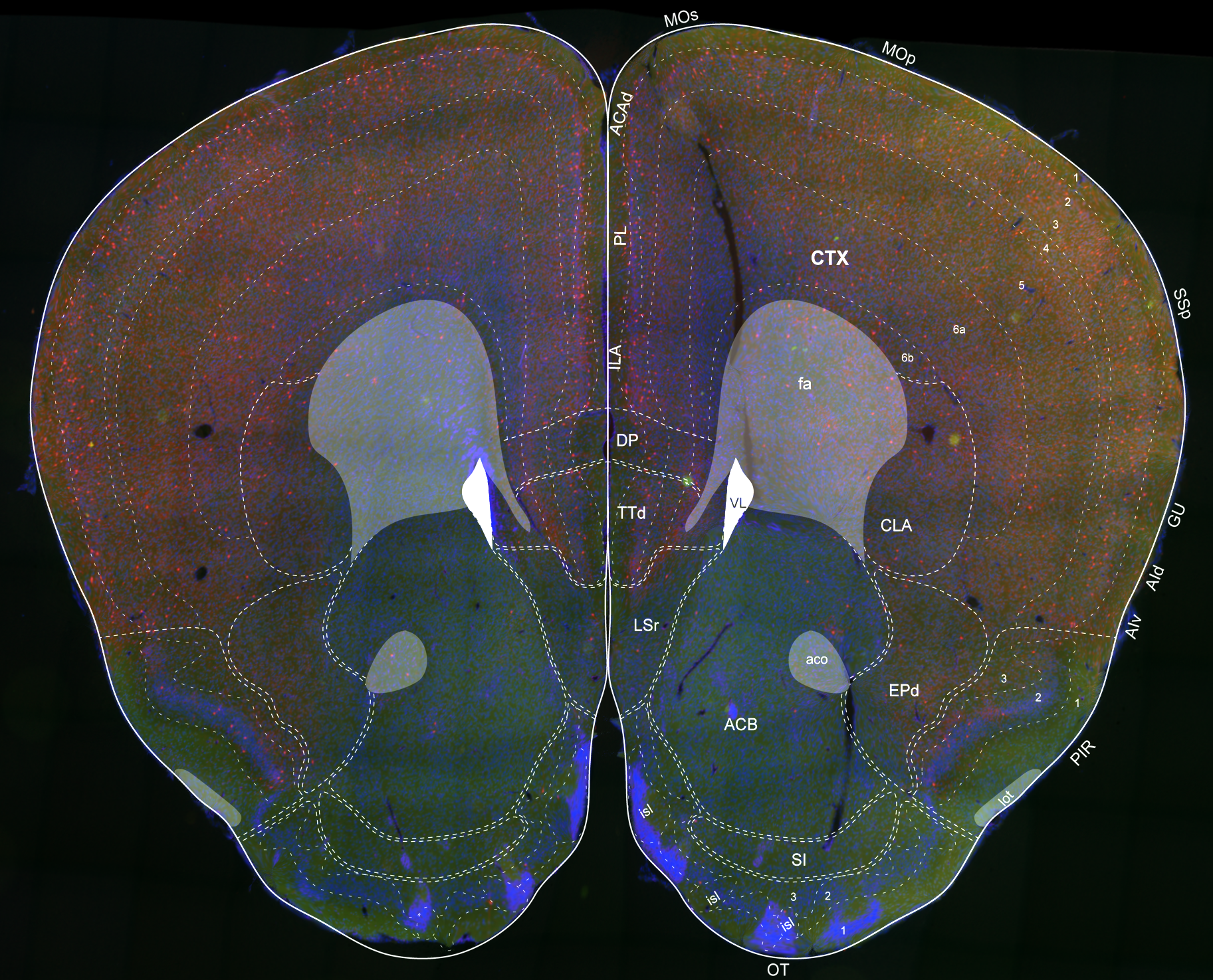
| Mouse Brain Model | Mesh Slicer | Registration |
 |
N. Agarwal, X. Xu, M. Gopi Geometry Processing of Conventionally Produced Mouse Brain Slice Images Journal of Neuroscience Methods, 2018 [arXiv Preprint] Supplementary Document [Bibtex] |
Related WorkN. Agarwal, X. Xu, M. Gopi Automatic Detection of Histological Artifacts in Mouse Brain Slice Images. In MICCAI, 2016. [PDF] |
Acknowledgements |
|
Project template was borrowed from Richard Zhang |